Dr. LeBlanc Recent Publications
Abstract: Cells maintain spatiotemporal control over biochemical processes through the formation and dissolution of biomolecular condensates, dynamic membraneless organelles formed via liquid–liquid phase separation. Composed primarily of proteins and nucleic acids, these condensates regulate key cellular functions, and their properties are influenced by the concentration and type of molecules involved. The structural versatility challenges the de novo design and assembly of condensates with predefined properties. Through feedback between computational and experimental approaches, we introduce a modular system for assembling condensates using nucleic acid nanotechnology. By utilizing programmable oligonucleotides and orthogonal synthesis methods, we control the structural parameters, responsive behavior, and immunorecognition of the products. Dissipative particle dynamics simulations predict some conditions to produce larger, well-defined condensates with compact, globular cores, while others result in smaller, more diffuse analogs. Fluorescence microscopy confirms these findings and microrheology demonstrates the viscoelastic adaptability of tested condensates. Nucleases trigger disruption of structures, and ethidium bromide intercalation protects condensates from digestion. Immunostimulatory assays suggest condensate-specific activation of the IRF pathway via cGAS-STING signaling. This study provides a framework for developing biomolecular condensates with customizable properties and immunorecognition for various biological applications.
- Time-resolved molecular mechanisms of essential biological processes. S LeBlanc. Multiscale Imaging and Spectroscopy V, PC128270O, 2024.
Abstract: My talk will focus on the application of single molecule time-resolved fluorescence spectroscopy as a tool to explore in real time the protein-nucleic acid interactions and dynamic structural rearrangements that drive biological pathways. We are currently studying the detailed mechanisms SARS-CoV-2 viral RNA processing, ribosome assembly, and DNA mismatch repair. I will present recent results from pulsed interleaved excitation fluorescence resonance energy transfer (PIE-FRET) and fluorescence correlation spectroscopy (FCS) experiments. We aim to completely characterize the impact of mutations linked to disease on the molecular function of enzymes to understand how complex diseases develop at the molecular level and to identify new therapeutic targets.
- Revealing pathways of ribosome assembly and viral RNA processing with time-resolved fluorescence. Sharonda LeBlanc, Lewis Alex Rolband, Kenya Gordon, Ben Clark, Jose Castaneda, Irene Silvernail, Andi Morgan, Patil Smruti, Meredith Frazier, Zoe Wright, Isha Wilson, Robin Evans Stanley. Biophysical Journal 123 (3), 25a, 2024.
Abstract: Large numbers of inanimate biomolecular machines organize across space and time and undergo drastic changes in shape to carry out complex biological processes. The detailed molecular mechanisms of these tightly regulated pathways are rarely known but driven by series of highly coordinated protein-protein and protein-nucleic acid interactions. Catching these transient events in action requires fast detection and high sensitivity, which is offered by time-resolved fluorescence methods. I will discuss single molecule time-resolved fluorescence microscopy and spectroscopy as a tool to explore in real time the interactions and structural rearrangements that drive specific pathways. We are currently studying the mechanisms of ribosome assembly, SARS-CoV-2 viral RNA processing, and DNA mismatch repair. We have designed assays for simultaneous Föster resonance energy transfer and fluorescence correlation spectroscopy measurements to investigate several fundamental questions. In my talk, I will present recent findings that reveal the dynamic interactions between essential enzymes, assembly factors, and nucleic acids. We aim to completely characterize the impact of mutations on the molecular function of enzymes to understand how complex diseases develop at the molecular level and to identify new therapeutic targets.
- A practical guide to time-resolved fluorescence microscopy and spectroscopy. Benjamin S Clark, Irene Silvernail, Kenya Gordon, Jose F Castaneda, Andi N Morgan, Lewis A Rolband, Sharonda J LeBlanc. bioRxiv, 2024.01. 25.577300
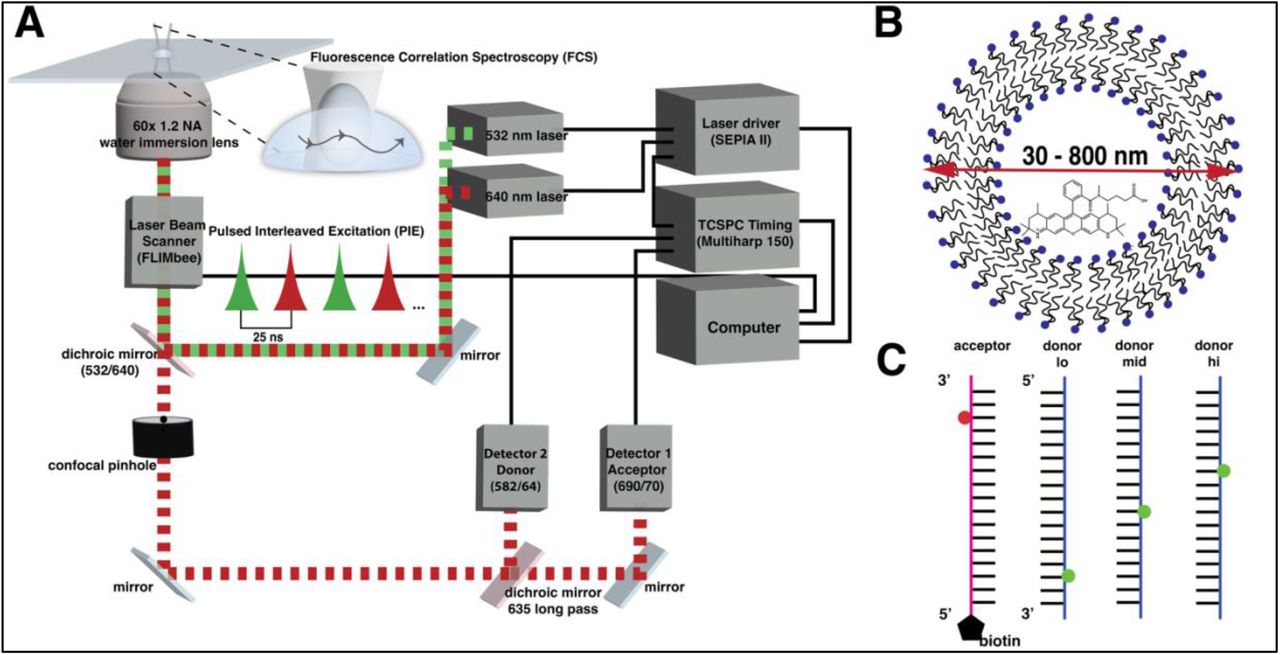
Abstract: Time-correlated single photon counting (TCSPC) coupled with confocal microscopy is a versatile biophysical tool that enables real-time monitoring of biomolecular dynamics across many timescales. With TCSPC, Fluorescence correlation spectroscopy (FCS) and pulsed interleaved excitation-Förster resonance energy transfer (PIE-FRET) are collected simultaneously on diffusing molecules to extract diffusion characteristics and proximity information. This article is a guide to calibrating FCS and PIE-FRET measurements with several biological samples including liposomes, streptavidin-coated quantum dots, proteins, and nucleic acids for reliable determination of diffusion coefficients and FRET efficiency. The FRET efficiency results are also compared to surface-attached single molecules using fluorescence lifetime imaging microscopy (FLIM-FRET). Combining the methods is a powerful approach to revealing mechanistic details of biological processes and pathways.
- Exploring cleavage activity of NSP15 using single molecule PIE-FRET. K Gordon, M Frazier, I Wilson, R Stanley, S LeBlanc. Biophysical Journal 122 (3), 72a, 2023.
Abstract: The NSP15 endoribonuclease is used by coronaviruses (CoVs) to avoid detection by host immune systems by cleaving viral RNA and therefore can be considered a possible therapeutic target for CoV diseases. Cryo-EM structures from prior studies have indicated NSP15 has a strong preference for cleaving uridine, but a more dynamic picture of enzyme kinetics would be useful for a complete understanding of enzyme activity. To explore real time cleavage dynamics of NSP15 in solution, we designed a single molecule assay using Pulsed Interleaved Excitation Fluorescence Resonance Energy Transfer (PIE-FRET) to monitor the cleavage of engineered fluorescently-labeled RNA substrates containing either A, U, or C at a specific site. We also used Fluorescence Correlation Spectroscopy (FCS) to determine diffusion times of RNA sub-populations during the cleavage process. To probe cleavage activity of individual complexes of NSP15 and RNA, we immobilized the RNA substrate and measured changes in the fluorescence intensity and lifetime over several minutes and performed FRET analysis. We found that PIE-FRET and FCS of the RNA substrates incubated with NSP15 suggest cleavage happens quickly for equimolar ratios of substrate and protein. Furthermore, analysis from both techniques indicates folding at the U base, which prior research has suggested may be a possible marker that host immune systems use for identifying viral RNA. Detailed analysis of the dynamics of the NSP15-RNA substrate interaction improves our understanding of NSP15 cleavage activity and may serve as a basis for exploring the kinetics of other enzymes implicated in human diseases.
- Investigating the impact of ATPase mutation on MutL function. A Drew, A Morgan, S LeBlanc. Biophysical Journal 122 (3), 331a, 2023.
Abstract: DNA mismatch repair (MMR) is an enzymatic pathway for fixing incorrect base pairing that may arise during DNA replication. Mutations in the enzyme MutL have been linked to Lynch syndrome, which is the leading cause of hereditary colorectal cancer. MutL is an important part of the MMR pathway and is thought to nick the error containing DNA strand for repair as well as perform other critical functions. To accomplish this, MutL has several large conformational changes that occur in the presence of ATP. While we believe these conformational changes are important for MMR, the exact order of the process and the direct effect of disease-related mutations are unknown. To better understand this process, we used Thermus Aquaticus MutL as a model system for human MutLalpha and mutated the ATPase domain by changing the glutamic acid in residue 28 to an alanine (E28A, aligned to E34A in human MLH1, a common mutation in Lynch syndrome patients). We then conducted an ATPase assay to compare wild-type and E28A activity and fluorescently labeled each MutL variant for single molecule fluorescence studies. We monitored MutL conformational dynamics under varying nucleotide conditions using pulsed interleaved excitation fluorescence resonance energy transfer (PIE-FRET) and fluorescence correlation spectroscopy (FCS) to reveal the impact of the E28A mutation on the dynamic function of MutL.
- Nucleotide dependent conformational changes of MutL. A Morgan, A Drew, A Atisa, MA Hinds, S LeBlanc. Biophysical Journal 122 (3), 41a, 2023.
Abstract: MutL is one of the enzymes required for mismatch repair (MMR). Mutations in MutL are linked to Lynch Syndrome, which predisposes individuals to colorectal and other cancers. In MMR, MutL is known to nick the error-containing daughter strand of mismatch DNA as well as form complexes with other MMR proteins such as MutS. MutL accomplishes this by undergoing a series of conformational changes which include extended, one-arm, semi-condensed, and condensed states in the presence of ATP. These dynamic conformational changes are needed to coordinate MMR, but the exact sequence of events and the impact of disease-associated mutations are unknown. We used site-directed mutagenesis to fluorescently label sites in the different domains of Thermus Aquaticus MutL, including the N-terminal, linker arms, and C-terminal domains. We expressed MutL variants with wild-type and ATPase-deficient activity and monitored their dynamic conformational changes under differing nucleotide conditions with pulsed interleaved excitation fluorescence resonance energy transfer (PIE-FRET) and fluorescence correlation spectroscopy (FCS) in solutions of freely diffusing molecules. Using the same technique, we also monitored the interactions of MutL with MutS, the enzyme that initially recognizes a mismatch, to reveal the specific interactions and conformational changes that are necessary to initiate mismatch repair.
